Miniaturized Sequencing Workflows for Microbiome and Metagenomic Studies
Authors: Jefferson Lai, Jared Bailey, Rabia Khan, and John Lesnick
Beckman Coulter Life Science, San Jose, CA, USA
Highlight
Our understanding of the role of the human microbiome in health and disease has grown rapidly in recent years. Amplicon sequencing of highly conserved 16S ribosomal RNA (rRNA) regions has long been the standard technique used to assess patient microbial diversity and is still heavily employed in academic and industrial institutions. As researchers ask more complex questions, with greater degrees of breadth and depth, they are turning to whole genome sequencing of the microbiome. Beyond that, microbial metatranscripts are also of interest, adding RNASeq to the techniques being used. For scientists looking to scale facilities and workflows to accommodate this growth field, the Echo® 525 Liquid Handler can be employed to miniaturize workflows, thus decreasing reagent cost and time spent per sample. Here, we introduce experiments of various sequencing workflows, miniaturizing them at least 10-fold. Our results show that miniaturizing sequencing workflows is possible and yields high quality data. The Echo 525 Liquid Handler enables the throughput needed for modern microbiome studies by reducing the time and cost per sample and processing them with speed and accuracy.
Miniaturizing Sequencing Workflows Saves Input, Reagent, and Time
16S rRNA amplicon sequencing is the prevalent technique used in phylogeny and medical microbiology, conferring many advantages over phenotypic methods. This workflow can yield the sample genus overnight, and with next-generation sequencing, thousands of samples can be analyzed, allowing for community-wide studies. We performed a standard Illumina 16S rRNA amplicon sequencing library preparation at miniaturized scale using the Echo 525 Liquid Handler. The samples were reference communities supplied by Zymo and ATCC. Detailed information can be found in Technical Note G123, Miniaturized 16S rRNA Amplicon Sequencing with the Echo Liquid Handler for Metagenomic and Microbiome Studies.
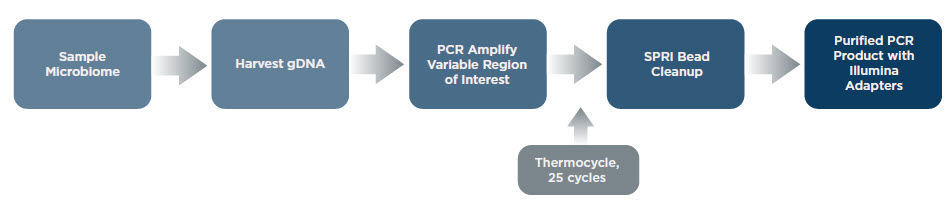
16S rRNA Amplicon Sequencing Workflow
Amplifying Sample Region of Interest and Adding Illumina Adapters
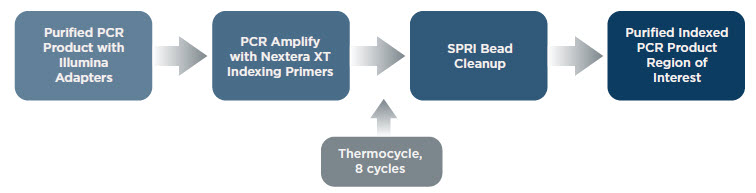
Attaching Nextera® XT Indices to Samples via Amplification from Adapters
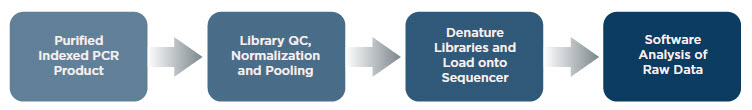
Library QC, Generating, and Analyzing Data
However, there are limitations to this method. 16S rRNA amplicon sequencing only captures prokaryotic diversity and misses eukaryotic and viral components of the microbiome. While additional amplicon sequencing of the internal transcribed spacer 1 (ITS1) region can capture fungal diversity, there is no known parallel technique for viral detection. Furthermore, these rRNA amplicon methods are generally only genusspecific. To obtain species-level differentiation, multiple variable regions of the rRNA need to be assessed in repeated experiments. Additionally, important strain information is not detected. Strain-to-strain variation is responsible for pathogenicity, toxins, virulence factors, epitopes, and antibiotic resistance characteristics.
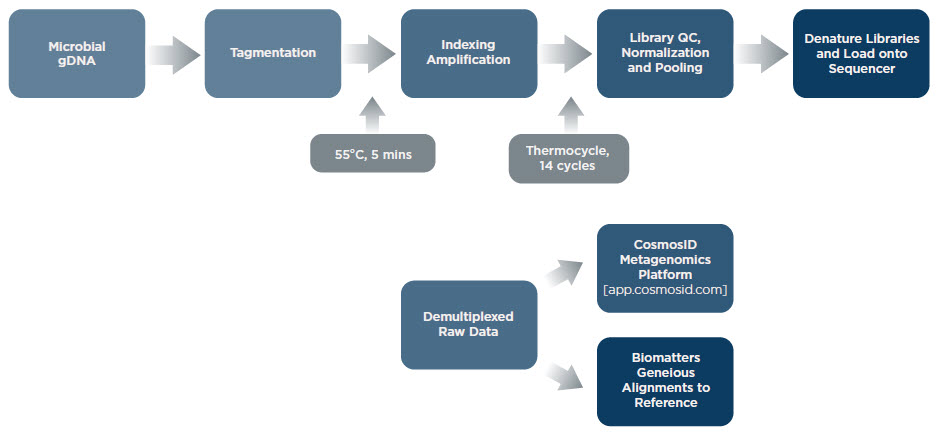
Whole genome sequencing (WGS) can determine strain-to-strain variation and has more applications beyond microbial community identification. Metagenomic homolog discovery from uncharacterized organisms is enabling pathway design for production of molecules of interest. Using WGS, biologists are prospecting for natural enzymatic solutions to their challenges. Miniaturizing WGS significantly lowers the cost to sequence and discover, as well as reducing process cycle time. We miniaturized the Illumina Nextera XT library preparation workflow for microbial whole genome sequencing. The samples were reference communities supplied by Zymo, ATCC, and NIST. Detailed information can be found in Technical Notes G121, Effective Miniaturization of Illumina Nextera XT Library Prep for Multiplexed Whole Genome Sequencing and Microbiome Applications, and G127, Whole Genome Sequencing of Microbial Communities for Scaling Microbiome and Metagenomic Studies.
Illumina Nextera® XT Library Preparation Workflow to Generate Libraries for Sequencing
Scientists also seek to understand the delicate and complex interplay of microbes within a community. Many ask questions that can be answered through metatranscriptomics. When large-scale community samples are studied, researchers can begin to draw connections between up-and down-regulation of genes in the presence and absence of other microbes or their byproducts. Our efforts to miniaturize RNASeq, typically involving reverse transcription of mRNA and Illumina Nextera XT library preparation of the resultant cDNA, are detailed in Technical Note G122, Echo® System-Enhanced SMART-Seq® v4 for RNA Sequencing.
Echo System-enabled Sequencing Workflows Yield Accurate and Reproducible Results
The Echo 525 Liquid Handler can be used to enhance QC measures. The instrument can accurately dispense lambda DNA for Picogreen standard curve in 25 nL increments, thus avoiding the need for serial dilution and potential propagation errors (Figure 1). The Echo 525 Liquid Handler is also used for fragment size analysis, dispensing samples and reagents quickly, enabling scalability in high-throughput studies (Figure 2). Lastly, the instrument possesses the unique ability to simultaneously normalize and pool hundreds of sequencing libraries in minutes.
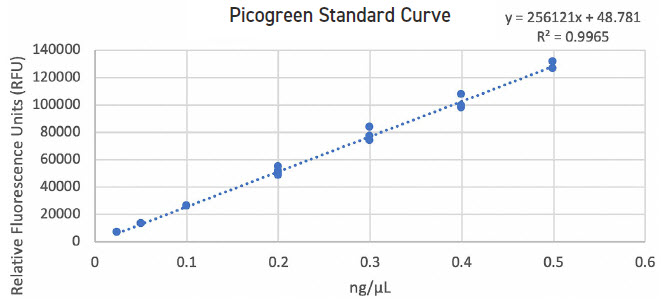
FIGURE 1: Picogreen standard curve generated by using the Echo 525 Liquid Handler in quadruplicate. The instrument is able to perform accurate and precise transfers of the lambda DNA used in this standard curve by direct dilution, avoiding propagation of errors in serial dilution.
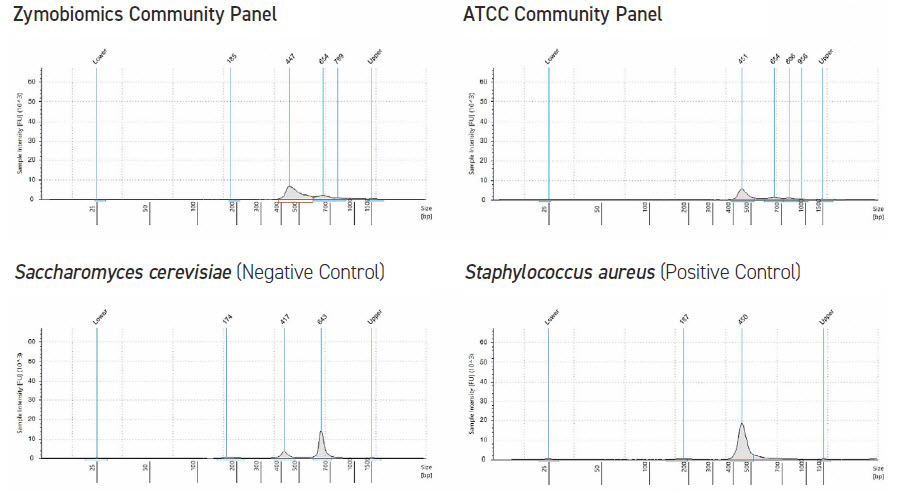
FIGURE 2: Agilent TapeStation 2200 electropherograms of 16S rRNA amplicon sequencing samples. We see the expected 450 base pair band for the positive control, a non-specific 650 base pair band for the negative (fungal) control, and that the reference communities contain both, as expected.
After sequencing of 16S rRNA amplicon libraries, a rarefaction curve was generated using CosmosID bioinformatic tools (FIGURE 3). This graph shows both Zymo and ATCC reference samples, replicated, and reaching alpha diversity saturation consistently. This graph demonstrates that a miniaturized process can produce more than enough reads to saturate the diversity of the samples. Higher complexity samples will require higher read depth, which can be accounted for during normalization and pooling with the Echo 525 Liquid Handler.
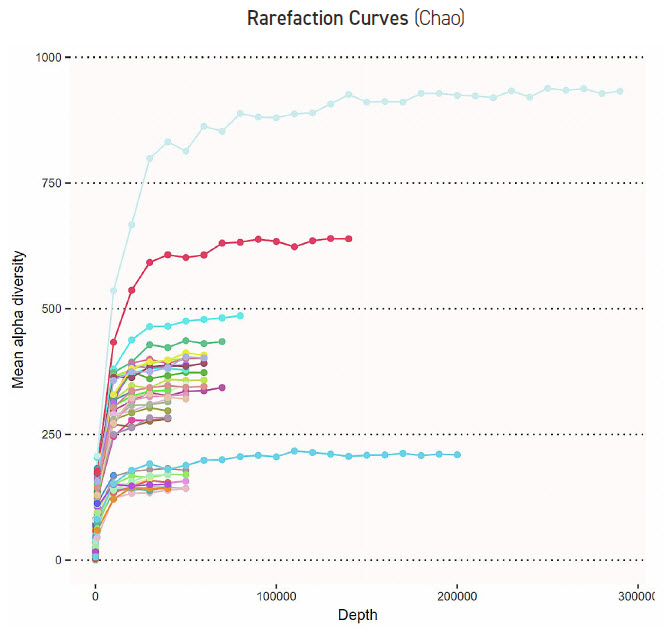
FIGURE 3: CosmosID bioinformatic chart. Rarefaction curve (Chao method) of 16S rRNA amplicon samples show that we consistently saturate alpha diversity for different communities, and we’ve generated adequate read depth in our miniaturized process.
After sequencing of whole genome Nextera XT prepared libraries, various analyses were run using CosmosID bioinformatic tools. When we compare replicates using a stacked bar graph with relative and logscale of diversity, we see very consistent results (Figure 4). Taking the replicate analysis one step further, we used a 3D principle component analysis graph to map the relationship between samples (Figure 5). We see that the miniaturized process can produce high quality and consistent sequencing libraries, which are in turn analyzed and clustered tightly together as expected.
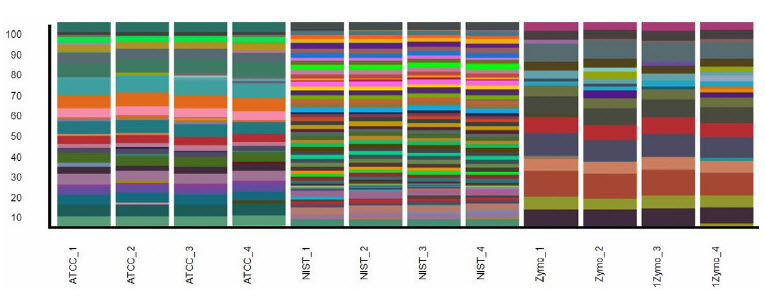
FIGURE 4: CosmosID bioinformatic chart. Stacked bar graph in relative and log-scale of sample diversity via whole genome sequencing. Using the Echo 525 Liquid Handler and Illumina Nextera XT, we see good consistency across all replicates.
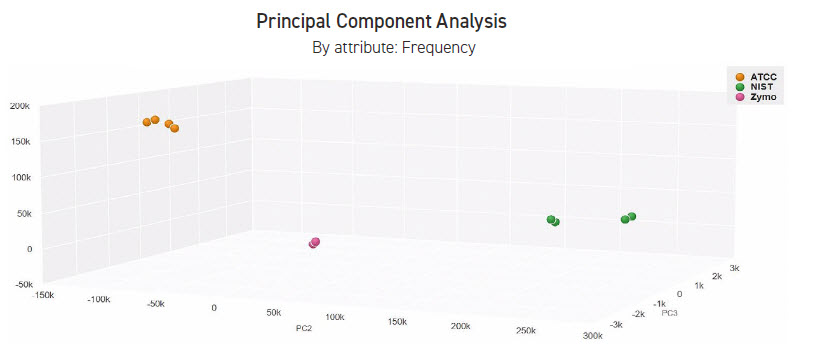
FIGURE 5: CosmosID bioinformatic chart. 3D Principle Component Analysis by frequencies via whole genome sequencing. Using the Echo 525 Liquid Handler and Illumina Nextera XT, we see good clustering and strong relationship between replicates of any given sample.
As researchers continue to deepen our understanding of the human microbiome, and as biologists explore the metagenomic space, our tools and analyses need to scale accordingly. A workflow utilizing the Echo 525 Liquid Handler can provide the solution to cost-effectively scale 16S rRNA and WGS of microbiomes to meet the demands of this new era of microbiome research.

